Research
Our overarching goal is to utilize high-throughput genomic and transcriptome data sets, predominantly based on Nanopore long-read sequencing, to investigate the role of DNA methylation in shaping RNA velocity trajectories across diverse biological systems and diseases. We hypothesize that DNA methylation influences gene expression regulation and reshapes RNA velocity trajectories. To address this hypothesis, we will develop innovative computational tools and leverage Nanopore long-read sequencing technology. These tools will enable us to accurately estimate RNA velocity and detect DNA methylation patterns, including rare or novel modifications.
Here are some themes and techniques that we currently work on:

We will develop a transformer-based machine learning tool for the direct detection of DNA methylation and evaluate its performance in diverse biological systems and diseases. Leveraging the signal-level information provided by Nanopore long-read sequencing, our novel transformer architecture will accurately detect DNA methylation patterns, including rare or novel modifications like 5hmC. The performance of our approach will be compared to existing methods for detecting DNA methylation using independent datasets.
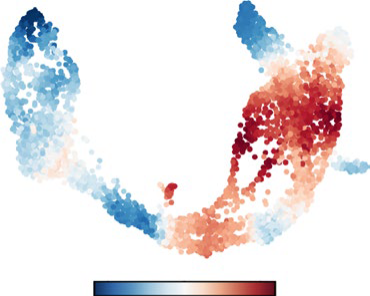
We will develop a computational toolbox that accurately estimates RNA velocity from Nanopore long-read RNA sequencing data. Leveraging the unique advantages of long-read sequencing, including rich signal-level information, our toolbox will model cell state dynamics based on RNA velocity, spliced/unspliced RNA abundances, latent times, and transcriptional states. The performance of our tool will be rigorously validated by comparing it to existing methods.
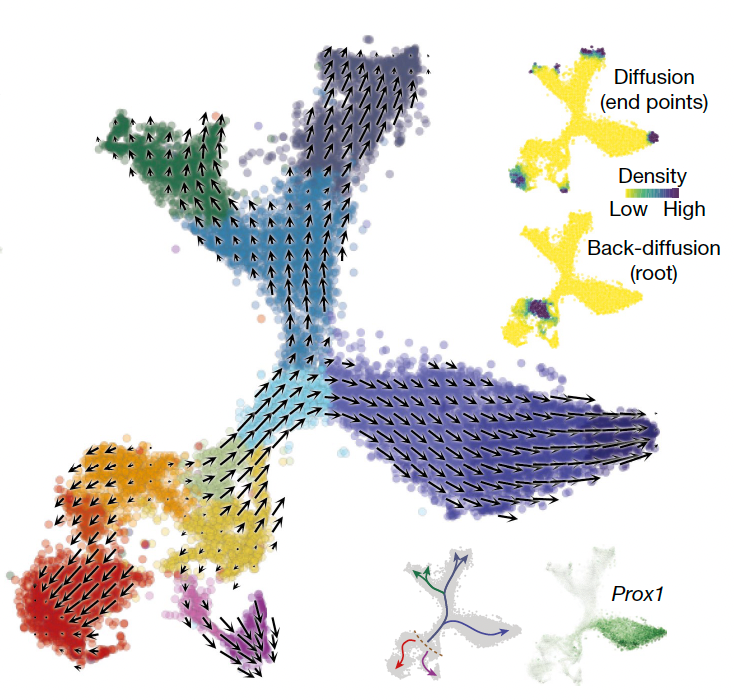
We will investigate the relationship between RNA velocity trajectories and DNA methylation patterns across diverse biological systems and diseases. Our computational tools will integrate RNA velocity and DNA methylation data to explore the interplay between these two regulatory mechanisms across different cell types and disease conditions.